| <meet.s(S-Plus)プログラムソース> |
| ダウンロード |
| # S-PLUS code for the detection of spatial
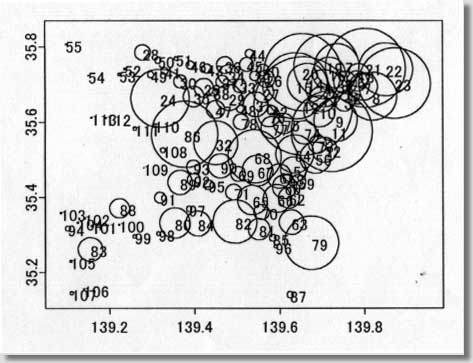
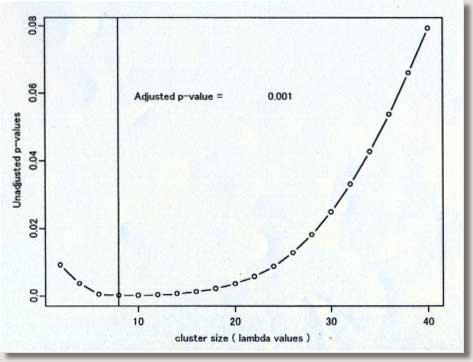
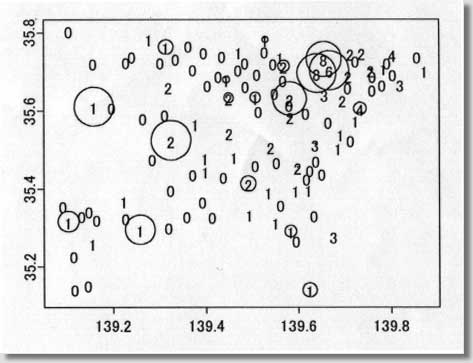
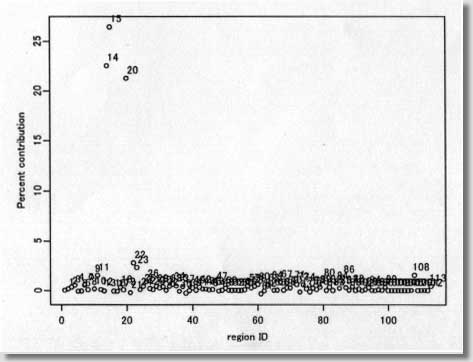
clustering program # called MEET (Maximized Excess Events Test) # # August 2000 (version 2.00) # # Programmed by # # Toshiro Tango # Division of Theoretical Epidemiology # Department of Epidemiology # The Institute of Public Health # 4-6-1 Shirokanedai, Minatoku, Tokyo, 108, Japan # E-mail: tango@iph.go.jp # # ---------------------------------------------------------------- # ( README FIRST ) # 1. Please read my paper carefully before using this program. # 2. You are free to use this program for noncommercial purposes # even to modify it but please cite this original program # # Reference: # Tango, T. A test for spatial disease clustering adjusted for # multiple testing. Statistics in Medicine 19, 191-204 (2000). # ----------------------------------------------------------------- # INPUT: # # nloop: The number of Monte Carlo replications, e.g., 999, 9999. # us$x: x-value of region centroid, e.g., longitude # us$y: y-value of region centroid, e.g., latitude # dc: distance matrix calculated from us$x and us$y # ( Depending on the user's situation, this should be modified !) # dat$j: j-th category of the confounders included (j=1,2,...,nk ) # dat$d: observed number of cases # dat$e: expected number of cases ( or population size) # kvec: a sequence of lambda values used for the profile p-values # # ------------------------------------------------------------------ # Major variables: # # nk: the number of strata or categories of all the confounders # included. For example, if age and sex are the confounders # included, with 18 different age groups, then there should be # 18x2 = 36 ( = nk ) categories. # nr: the number of regions # nn: the number of cases # kkk: the number of lambda values # pxx: sorted p-values due to Monte Carlo replications under # the null # ss : per cent contribution to C (equation 14) # top: region id sorted in the order of the magnitude of ss # (e.g., top[1] denote the most likely location of cluster, # if any.) # pres: The resultant adjusted p-value based on Monte Carlo # replicates # # +++++ An example of execution ++++++++++++++++++++++++++++++++++++ # # 100 cases: the random sample from clustering model A shown in # Figure 2 of my paper. The result will be the one # similar to Figure 3 and 4. The difference will be # due to Monte Carlo replicates produced. # These data files are added at the end of email. # # nloop<-999 nk<- 1 kvec<- c(1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20) kvec<- kvec*2 us <-scan("d:/demo/kanto.geo",list(id=0,x=0,y=0)) dat <-scan("d:/demo/kanto.dat",list(id=0,j=0,d=0,e=0)) # # These three data files are added at the end of this file. # It takes about 13 minutes using IBM thinkpad 600E. # # ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++ par(mfrow=c(1,1)) start<-date() nr<-length(us$id) qdat<-rep(0,nr) p<-rep(0,nr) w<-matrix(0,nr,nr) nn<-sum(dat$d) kkk<- length(kvec) dc<-matrix(0,nr,nr) regid<-us$id frq<-matrix(0,nloop,nr) pval<-matrix(0,nloop,kkk) pxx<-rep(0,nloop) pcc<-pxx for (i in 1:nr){ for (j in i:nr){ dij<-sqrt(((us$x[i]-us$x[j])*90.15)**2 + # An example ((us$y[i]-us$y[j])*110.9)**2) # for Tokyo area # dij<-sqrt(((us$x[i]-us$x[j]))**2 + # ((us$y[i]-us$y[j]))**2) dc[j,i]<- dij dc[i,j]<- dij } } # # Calculation of (1) null distribution of test statistic Pmin # for (j in 1:nk){ jdat<- dat$d[dat$j == j] nj<-sum(jdat) poo<-dat$e[dat$j == j] p1<-poo/sum(poo) pp<-matrix(p1) w1<-diag(p1)-pp%*%t(pp) qdat<- qdat + jdat p<- p+nj*p1 w<- w+nj*w1 py<-p1 vv<-py/sum(py) cvv<-cumsum(vv) for (ijk in 1:nloop) { r1<-hist(runif(nj),breaks=c(0,cvv),plot=F) frq[ijk,]<- frq[ijk,] + r1$count } } frq<-frq/nn ori<- qdat qdat<-qdat/nn w<- w/nn p<-p/nn symbols(us$x,us$y,circles=p,inch=0.50) # Figure 1 text(us$x,us$y,regid,adj= -0.5, col=8) # for (k in 1:kkk){ rad<- kvec[k] ac <- exp( -4 * (dc/rad)^2 ) hh <- ac%*%w hh2<- hh%*%hh av <- sum(diag( hh )) av2<- sum(diag( hh2 )) av3<- sum(diag( hh2%*%hh )) skew1<- 2*sqrt(2)*av3/ (av2)^1.5 df1<- 8/skew1/skew1 eg1<-av vg1<-2*av2 for (ijk in 1:nloop){ q<-frq[ijk,] gt<-(q-p)%*%ac%*%(q-p)*nn stat1<-(gt-eg1)/sqrt(vg1) aprox<-df1+sqrt(2*df1)*stat1 pval[ijk,k]<- 1-pchisq(aprox, df1) } } for (ijk in 1:nloop){ pcc[ijk]<-min( pval[ijk,] ) } pxx<- sort( pcc ) # # Calculation of (2) test statistic Pmin and its adjusted p-value # of observed data # ptan<-rep(0,kkk) gtt<-rep(0,kkk) jjj<-1:kkk ep<-p*nn po<-ppois(ori,ep) pa<-(po-0.90)/0.10 p1<-ifelse(po > 0.9 & ori>0, pa^3, 0) symbols(us$x,us$y,circle=p1,inch=0.2) # Figure 2 text(us$x, us$y, ori, col=8) par(cex=0.5) par(mar=c(4,3,3,5)) par(mfrow=c(1,2)) par(mgp=c(1.5,0.5,0)) # for (k in 1:kkk) { ti<-date() rad<- kvec[k] ac <- exp( -4 * (dc/rad)^2 ) hh <- ac%*%w hh2<- hh%*%hh av <- sum(diag( hh )) av2<- sum(diag( hh2 )) av3<- sum(diag( hh2%*%hh )) skew1<- 2*sqrt(2)*av3/ (av2)^1.5 df1<- 8/skew1/skew1 eg1<-av vg1<-2*av2 gt<-(qdat-p)%*%ac%*%(qdat-p)*nn gtt[k]<-gt stat1<-(gt-eg1)/sqrt(vg1) aprox<-df1+sqrt(2*df1)*stat1 ptan[k]<- 1-pchisq(aprox, df1) } # # Figure 3 p22<-min( ptan ) at<-c(pxx,p22) pres<-length(at[at<=p22])/(length(pxx)+1) pind<-jjj[ptan==min(ptan)] plot(kvec,ptan,pch=1,type="b",xlab="cluster size ( lambda values ) ", ylab=" Unadjusted p-values") abline(v=kvec[pind]) text((max(kvec)+2*min(kvec))/3,(3*max(ptan)+min(ptan))/4, "Adjusted p-value = ",col=8) text((1.5*max(kvec)+min(kvec))/2.5,(3*max(ptan)+min(ptan))/4, pres,col=8) # # Figure 4 rad<- kvec[pind] ac <- exp( -4 * (dc/rad)^2 ) ss<-ac%*%(qdat-p)*nn ss<-(qdat-p)*ss ss2<-ss ss<-ss/sum(ss)*100 plot(regid,ss,pch=1,xlab=" region ID ", ylab=" Percent contribution ") text(regid+2,ss+1,regid,col=8) top<-regid[sort.list(-ss)] end<- date() # # End of S-plus file # ---------------------------------------------------------------- # Below are two data sets for example run # # example data : kanto.geo (id, longitude, latitude) # 1 139.755 35.69056 2 139.7753 35.6675 3 139.7544 35.655 4 139.7075 35.68972 5 139.7564 35.70472 6 139.7844 35.70889 7 139.7989 35.69472 8 139.8197 35.66833 9 139.7333 35.60583 10 139.6956 35.62889 11 139.7236 35.57611 12 139.6564 35.64278 13 139.7014 35.66083 14 139.6669 35.70361 15 139.6397 35.69639 16 139.7186 35.72917 17 139.7369 35.74944 18 139.7867 35.72444 19 139.7122 35.74778 20 139.6553 35.7325 21 139.8036 35.74389 22 139.8508 35.74 23 139.8717 35.70333 24 139.3192 35.66333 25 139.4225 35.69083 26 139.5694 35.71472 27 139.5625 35.68 28 139.2781 35.785 29 139.4808 35.66556 30 139.3667 35.70806 31 139.5442 35.6475 32 139.45 35.54528 33 139.5064 35.69611 34 139.4806 35.72528 35 139.3983 35.66806 36 139.4717 35.75139 37 139.4656 35.70778 38 139.4444 35.68056 39 139.5417 35.7225 40 139.5622 35.73833 41 139.33 35.73556 42 139.5819 35.63167 43 139.4297 35.74222 44 139.5297 35.7825 45 139.5231 35.75306 46 139.3906 35.75167 47 139.4497 35.63389 48 139.5078 35.63472 49 139.2969 35.72611 50 139.3142 35.76417 51 139.3572 35.76889 52 139.2361 35.74139 53 139.2233 35.72667 54 139.1519 35.72361 55 139.0994 35.80639 56 139.6856 35.50556 57 139.6328 35.47389 58 139.6197 35.45056 59 139.6453 35.44139 60 139.6119 35.42833 61 139.5992 35.45667 62 139.6219 35.39917 63 139.6278 35.33417 64 139.6364 35.51583 65 139.5358 35.39306 66 139.5944 35.39722 67 139.5481 35.47139 68 139.5411 35.50917 69 139.5022 35.46306 70 139.5569 35.36139 71 139.4917 35.41444 72 139.7072 35.52639 73 139.6906 35.54056 74 139.6592 35.57306 75 139.6108 35.59639 76 139.5756 35.61694 77 139.5825 35.58611 78 139.5094 35.60083 79 139.6753 35.27889 80 139.3531 35.33222 81 139.5511 35.31528 82 139.4947 35.33583 83 139.1556 35.26139 84 139.4078 35.33056 85 139.5833 35.29222 86 139.3764 35.56806 87 139.6239 35.14083 88 139.2233 35.37139 89 139.3656 35.44 90 139.4611 35.48417 91 139.3183 35.39944 92 139.3986 35.44694 93 139.3975 35.48111 94 139.1031 35.3175 95 139.4325 35.43361 96 139.59 35.26917 97 139.3869 35.36972 98 139.3144 35.30361 99 139.2581 35.29639 100 139.2219 35.3275 101 139.1594 35.32361 102 139.1425 35.345 103 139.0864 35.35917 104 139.1264 35.33306 105 139.1097 35.22944 106 139.1406 35.155 107 139.1114 35.14472 108 139.3247 35.52583 109 139.2797 35.47889 110 139.3061 35.5925 111 139.2594 35.58306 112 139.1922 35.61111 113 139.1586 35.61167 # # example data: kanto.dat (id, stratum no, number of cases, population) # 1 1 0 39472 2 1 0 68041 3 1 0 158499 4 1 2 296790 5 1 2 181269 6 1 1 162969 7 1 0 222944 8 1 3 385159 9 1 4 344611 10 1 2 251222 11 1 1 647914 12 1 3 789051 13 1 0 205625 14 1 6 319687 15 1 8 529485 16 1 0 261870 17 1 2 354647 18 1 0 184809 19 1 2 518943 20 1 8 618663 21 1 4 631163 22 1 0 424801 23 1 1 565939 24 1 2 466347 25 1 0 152824 26 1 2 139077 27 1 0 165564 28 1 1 125960 29 1 0 209396 30 1 0 105372 31 1 0 197677 32 1 2 349050 33 1 0 105899 34 1 0 164013 35 1 0 165928 36 1 1 134002 37 1 0 100982 38 1 1 65833 39 1 0 75144 40 1 1 95146 41 1 0 58062 42 1 2 74189 43 1 0 75132 44 1 1 67539 45 1 0 113818 46 1 0 65562 47 1 2 144489 48 1 1 58635 49 1 0 50387 50 1 1 52103 51 1 0 30967 52 1 0 16444 53 1 0 21553 54 1 0 3808 55 1 0 8752 56 1 1 250100 57 1 0 205510 58 1 0 76978 59 1 0 116634 60 1 0 194617 61 1 2 195795 62 1 1 168846 63 1 0 197753 64 1 3 305774 65 1 1 238536 66 1 1 224036 67 1 0 248882 68 1 2 426663 69 1 0 119575 70 1 0 123766 71 1 2 126866 72 1 0 200056 73 1 1 142320 74 1 0 187707 75 1 0 165081 76 1 0 175570 77 1 2 177742 78 1 0 125127 79 1 3 433358 80 1 0 245950 81 1 1 174307 82 1 1 350330 83 1 1 193417 84 1 0 201675 85 1 1 56704 86 1 1 531542 87 1 1 52440 88 1 1 155620 89 1 0 197283 90 1 1 194866 91 1 0 89567 92 1 1 105822 93 1 1 112102 94 1 1 42600 95 1 0 77926 96 1 0 29536 97 1 0 44532 98 1 0 31599 99 1 1 29415 100 1 0 10054 101 1 0 14895 102 1 0 13097 103 1 0 14342 104 1 0 11941 105 1 0 19365 106 1 0 9588 107 1 0 27717 108 1 2 40424 109 1 0 3549 110 1 0 21535 111 1 0 28038 112 1 0 10592 113 1 1 10729 # -------------- End of all files ---------------------- |
| <プログラム処理結果サンプル> |
 |
 |
 |
 |
